RStudio Server
Users can launch an RStudio Server instance on Palmetto through Open OnDemand. RStudio Server provides a web based RStudio environment.
Starting a RStudio Server
-
Go to the OpenOD website.
-
Log in with your Palmetto user ID and password:
-
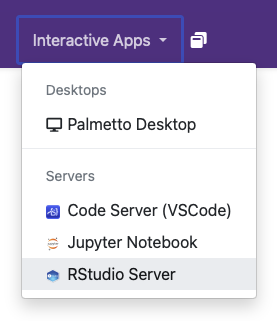
Once you are logged in, click on Interactive Apps on the top navigation bar.
-
Click on the RStudio Server link.
-
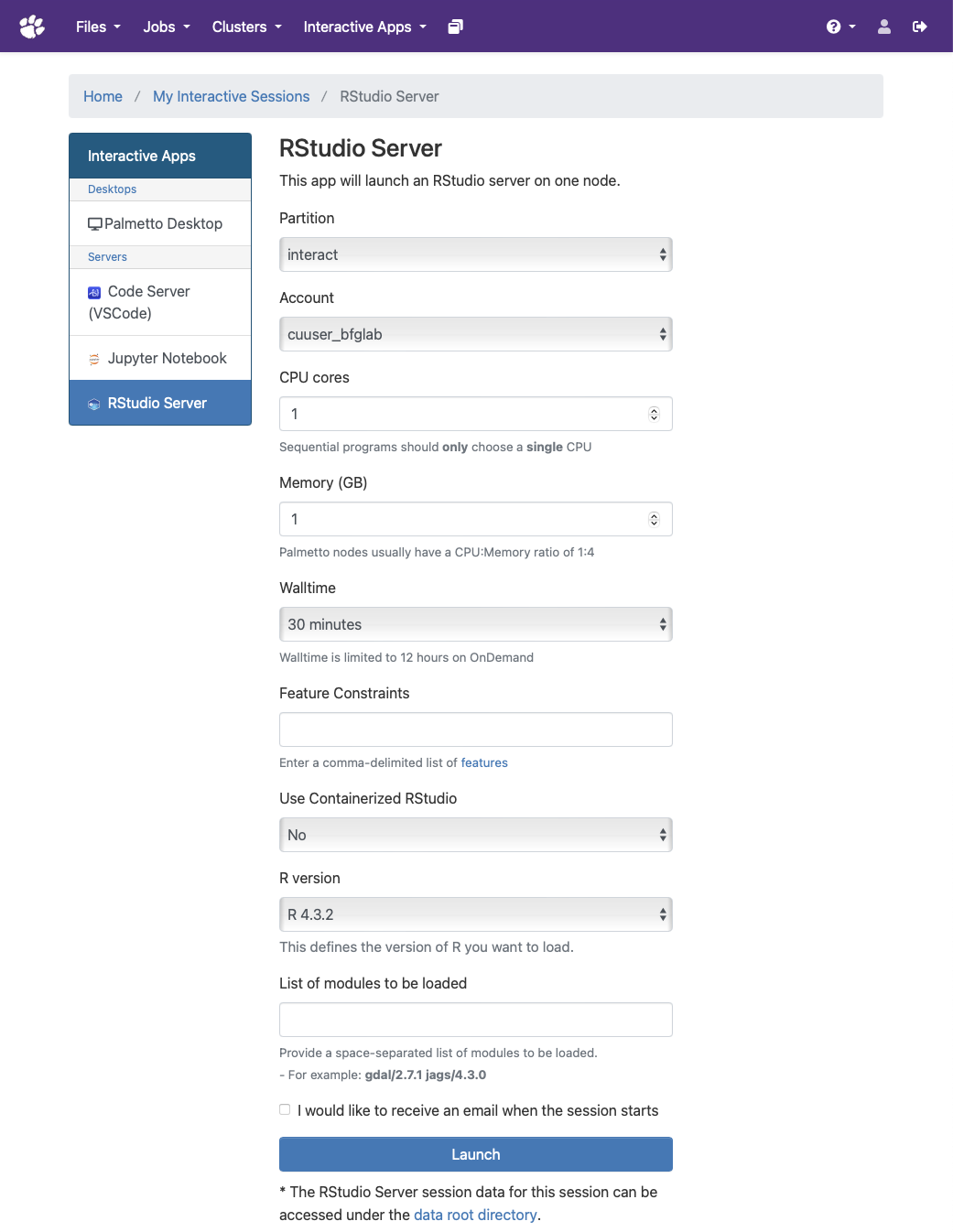
Select the resources (CPU cores, memory, walltime, etc.) required for your session.
Some R packages may require certain modules loaded. Be sure to include them in the "List of modules to be loaded". Some guidance is available below for module requirements for certain popular packages.
infoYou must specify an exact module name and version that is available on our cluster. To determine the exact name and version, use the
module availcommand over an ssh session. If you specify a module that does not exist, no modules will be loaded.cautionChanging the "List of modules to be loaded" in the resource selection form when launching RStudio may not immediately take effect if RStudio restores a previous session. To force the new modules to be available to RStudio, select Session->Restart R once RStudio is running.
-
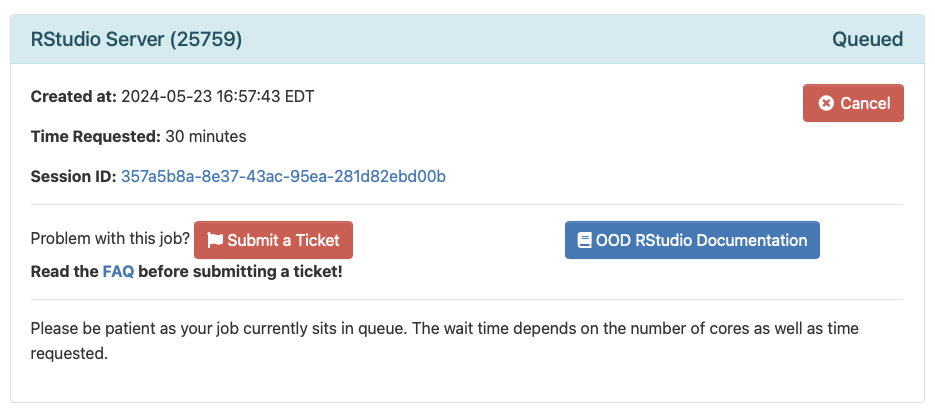
Once you launch your server you will be taken to your current list of interactive sessions. Your job will be queued until resources are available to handle your request.
-
Once your server has had it's resources allocated you can connect to your server with a button that says
Connect to RStudio Server, your browser will then be sent to the RStudio interface.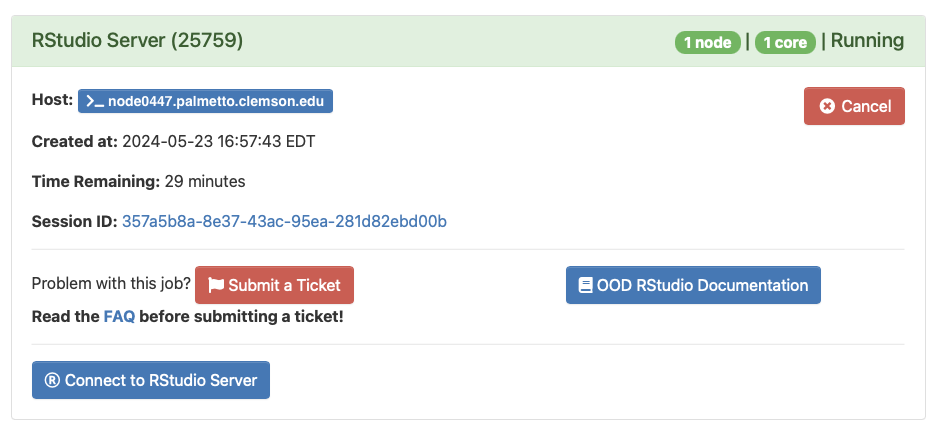
Installing Popular R Packages
Installing most packages in your RStudio server is as simple as running:
install.packages("package name")
These packages will install in your home directory and will be available for future RStudio sessions you run.
Some R packages require system libraries that may require loading libraries
before installing. To check to see if this is needed, search the
CRAN website
for your package. If it lists SystemRequirement, these are system level
libraries that must be loaded before the R package can be installed. For
example, sf requires proj, gdal,
geos, and sqlite3. These are already installed in the cluster as
modules so you must
add them in the "List of modules to be loaded" field
before launching the R Studio Server in Open OnDemand. More examples and
Palmetto cluster specific instructions for popular R packages follow.
terra and sf
Required modules:
gdal/3.8.3 geos/3.12.1 sqlite/3.43.2
Although the sqlite module is loaded, the configure scripts for terra and
sf fail to find it. To install, you'll need to manually pass a configure
argument:
install.packages('terra', configure.args=c('--with-sqlite3-lib=/software/slurm/spackages/linux-rocky8-x86_64/gcc-12.3.0/sqlite-3.43.2-brfrunmqjf77zexoe7umsc3wv5hcb5op/lib'))
install.packages('sf', configure.args=c('--with-sqlite3-lib=/software/slurm/spackages/linux-rocky8-x86_64/gcc-12.3.0/sqlite-3.43.2-brfrunmqjf77zexoe7umsc3wv5hcb5op/lib'))
If the operation fails the first time, try again. There have been instances of it not working at first, but working on the second install attempt.
lwgeom
Required modules:
gdal/3.8.3 geos/3.12.1 sqlite/3.43.2
The sf package is a dependency for lwgeom and you may want
to install it first.
Although the sqlite module is loaded, the configure script for lwgeom fails
to find it. To install, you'll need to manually pass a configure argument.
install.packages("lwgeom", configure.vars=c("INLIBS='-L/software/slurm/spackages/linux-rocky8-x86_64/gcc-12.3.0/sqlite-3.43.2-brfrunmqjf77zexoe7umsc3wv5hcb5op/lib'"))
Rmpfr (required for many other packages like corHmm)
Required modules:
mpfr/4.2.0
The Rmpfr package cannot seem to find our cluster install of mpfr without
some help. Add the following lines to the file ~/.R/Makevars (create the file
and directory if needed):
LDFLAGS=-L/software/slurm/spackages/linux-rocky8-x86_64/gcc-12.3.0/mpfr-4.2.0-qqprljzv7j6tdvxu7jmshqrzbvioh73w/lib
CFLAGS=-I/software/slurm/spackages/linux-rocky8-x86_64/gcc-12.3.0/mpfr-4.2.0-qqprljzv7j6tdvxu7jmshqrzbvioh73w/include
Now the standard install should work:
install.packages("Rmpfr")